Overview
Below you can find more information on specific projects, including links to videos, tutorials, webapps and code. For more up to date information you can also have a look at our github repository.

Our team is pioneering the usage of Virtual Reality technology for interactively exploring large networks and other complex datasets. You can find a series of introductory videos on a dedicated website. In 2021 we published our first proof-of-concept of a VR platform for biological network analysis, you can read the paper here. Since then, we have been constantly updating and extending our platform. Detailed instructions and tutorials can be found on our github page.
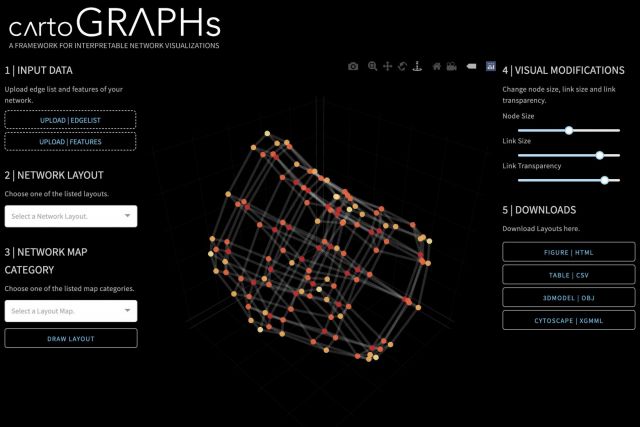
Networks offer a powerful visual representation of complex systems. In 2022, we published CartoGraphs, a framework for generating a diverse set of network layouts for highlighting and visually inspecting chosen characteristics of a network. The resulting visualizations are interpretable and can be used to explore complex datasets, such as large-scale biological networks. A general introduction to CartoGraphs can be found on a dedicated website and a research briefing. Our webapp allows users to generate their own CartoGraphs, all source code and detailed instructions are available on github.
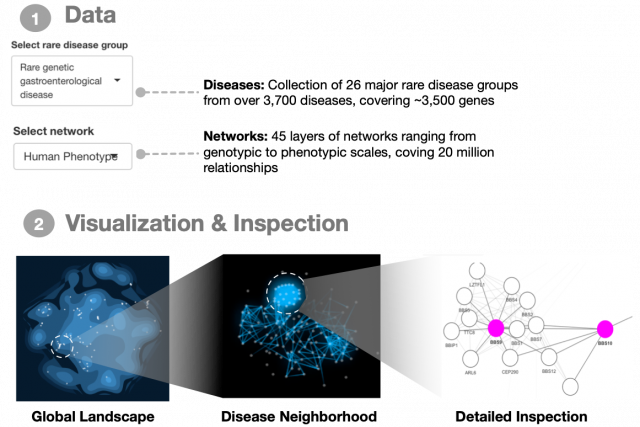
Rare genetic diseases affect various levels of biological organization between the genome and the whole organism. You can find a general introduction on this topic here. In 2021, we published a comprehensive analysis of 3,771 rare diseases and a their connection patterns on a multiplex network consisting of over 20 million gene relationships that are organized into 46 network layers spanning six major biological scales between genotype and phenotype. You can explore the results and investigate the connections among genes associated to a disease of interest in our interactive webapp. The code for all analyses presented in the paper can be found on our github page. The repository also contains a walk-through guide for reproducing the figures and analyses.
We have developed a series of tools for analyzing microscopy images from high-content imaging screens. In 2021, we published a dedicated package for the high-performance programming language Julia. The package allows for performing robust multidimensional profiling and comes with data structures and helper functions especially designed for high-content imaging-based morphological profiling. You can access the package from the Julia repository or from github.
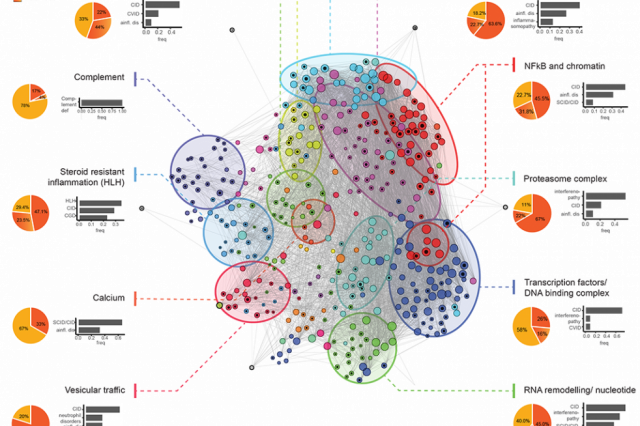
In a project that is currently under review for publication, we developed a network-based framework for integrating all currently known monogenic immune defects and their molecular interactions. We identify the AutoCore, that is, the set of genes and their molecular interactions that are essential for immune homeostasis. A brief overview of the project can be found on a dedicated website. We implemented a webapp where the AutoCore can be interactively explored. The code is available on github.