The human body contains over 200 different cell types, varying in size, shape, and function. The differentiation and subsequent maintenance of these distinct cell types are governed by complex gene regulatory networks that orchestrate the dynamic activation and deactivation of genes. Abnormalities in these networks may lead to dysfunctional expression programs: for example, uncontrolled cell proliferation. In order to understand the conditions resulting in disease, we must understand the underlying gene regulatory networks governing the gene expression program.
Gene regulatory network remodeling in cell differentiation trajectories
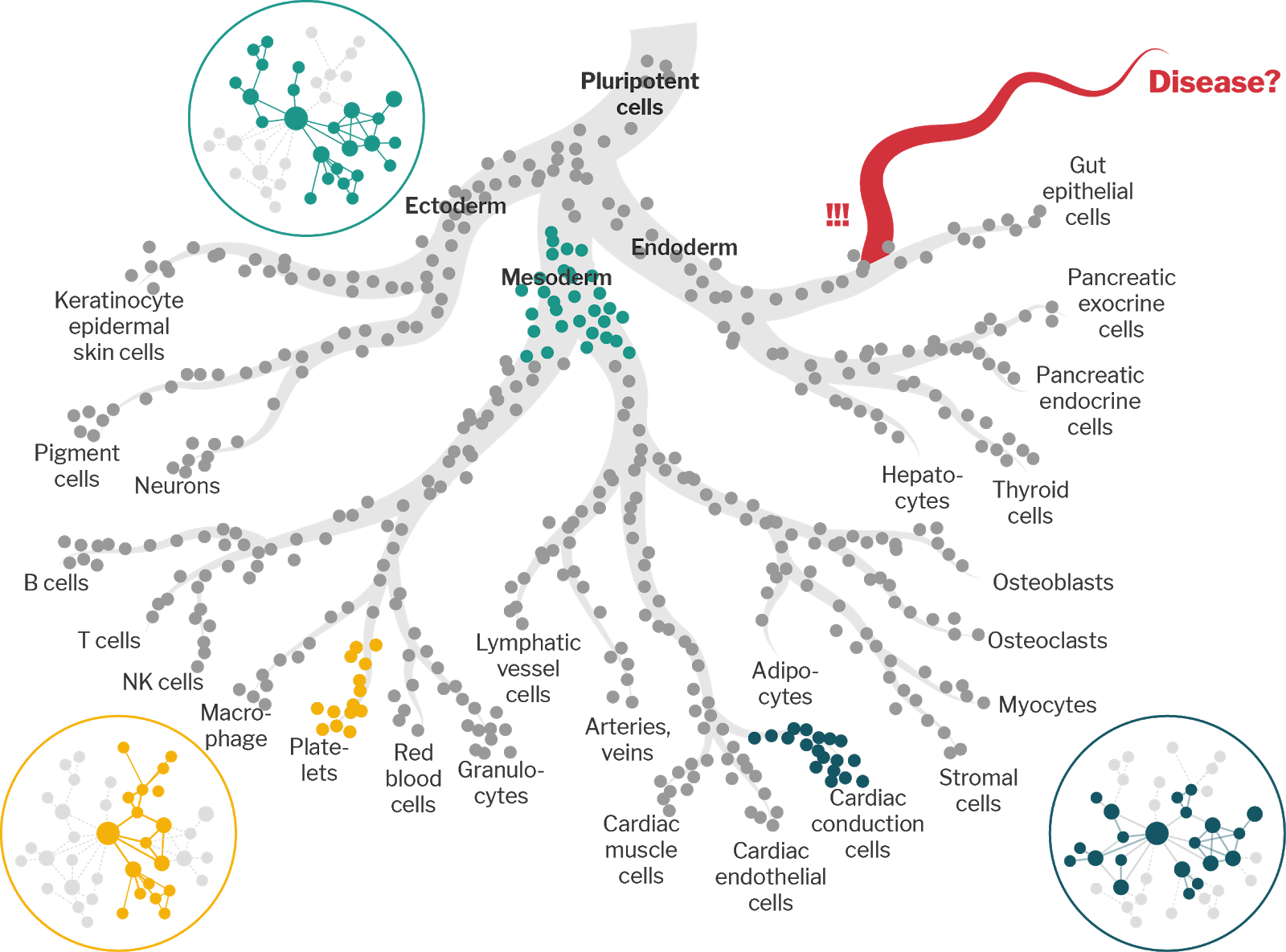
As cells move through the differentiation space, the networks that govern gene regulation are remodeled in order to achieve the appropriate gene expression program. While statistical physics and network theory have demonstrated numerous relationships between the structure of networks and the dynamic processes that act on them, few studies link these mathematically rigorous principles to gene regulatory networks, none at the level of cell-trajectory-states. We hypothesize that the gene regulatory networks of different cell-trajectory-states along the differentiation trajectory – e.g. transitory, branching, or terminal states – are each characterized by distinct structural features.
The overall goal of this project is to understand the fundamental architecture of gene regulatory networks associated with cell differentiation processes. In particular, our investigation of how gene regulatory networks are remodeled through differentiation processes will reveal basic principles relating topological network architecture to the dynamics possible along the epigenetic landscape. These basic principles can lead to insights applicable to other processes of differentiation, reprogramming, and disease.