Biomolecules exert their functions through interactions with each other. In analogy to the genome representing a blueprint for the assembly of the molecular parts of biological systems, the interactome of all their interactions can be understood as a blueprint for their collective functions. Our current knowledge of the human interactome comprises almost 20k proteins connected by more than 300k physical interactions. Over the last two decades, numerous relationships have been revealed between topological and functional interactome characteristics, allowing us to interpret the interactome as a map to systematically investigate both healthy and disease associated processes using network-based approaches. At the core of such approaches lie notions of locality, distance and neighborhoods within the network. Their most straightforward definitions that only rely on direct connectivity suffer from several conceptual and methodological limitations: (i) The vast heterogeneity among the degrees within the interactome translates immediately into a vast heterogeneity of neighborhood sizes: While immediate neighbors of hubs can cover more than 10% of all nodes in the network, the majority of nodes have only a few connections. (ii) Neighborhoods defined by nearest neighbors do not take connections between them into account.
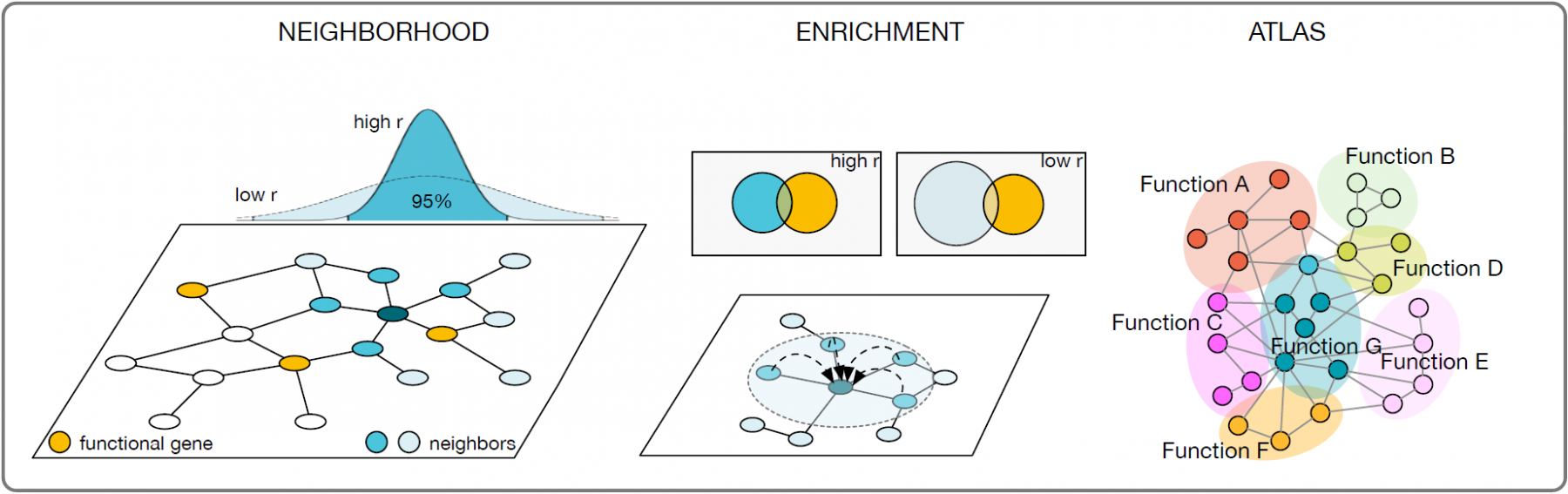
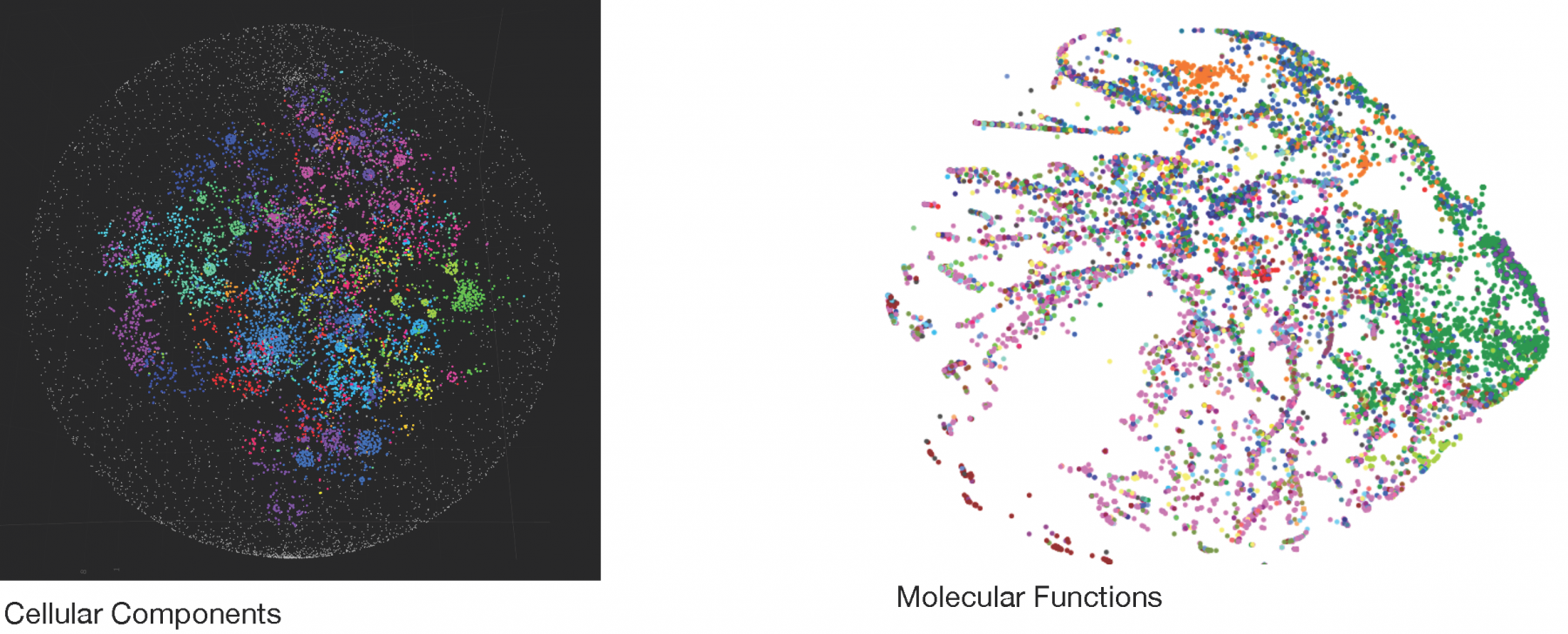
Here, we present a novel approach for defining and systematically exploring local neighborhoods in interactome networks using a diffusion process. We describe how the dynamic process incorporates local structural properties around individual nodes and demonstrate that it can balance out large neighborhoods around hubs, while giving low degree degree nodes more prominence. The resulting neighborhoods can be functionally enriched using biological annotations, which can be used to annotate individual genes or gene sets. Finally, we apply our methodology to construct a global functional atlas of the interactome.
Network informed functional enrichment for biological networks